DNAnexus
Workflows written in the WDL format can easily be imported into the DNAnexus platform using tools provided by DNAnexus: dx-toolkit and dxCompiler. Below demonstrates how to run the 02_download_collection_files.wdl workflow via DNAnexus. This workflow should take between 5-10 minutes to run.
Prerequisites
Before beginning, ensure you have the following:
- A DNAnexus platform account (including free tier)
- java 8+
- python 3.5+
- (Optional) cURL
- (Optional) git
Getting Started
The following software and workflows are required and will be installed and used as part of this tutorial:
Download the worked examples repository by one of the following methods.
- Clone with Git
1git clone https://github.com/DNAstack/dnastack-client-library-worked-examples.git - Or download and unzip
It is assumed that all commands will be run from the root directory of this repository on your local machine. From the directory where you’ve downloaded the examples repository:
| |
Download the latest version of dxCompiler by one of the following methods. This allows direct conversion and uploading of WDL workflows to DNAnexus.
- Using cURL:
1curl -o dxCompiler.jar -L https://github.com/dnanexus/dxCompiler/releases/download/2.4.6/dxCompiler-2.4.6.jar - Or download from the releases page to the current directory and rename the file to
dxCompiler.jar.
Download and initialize dx-toolkit. You will be prompted to log into your DNAnexus account.
| |
Create a project on DNAnexus where the workflow and associated files will be stored.
Running the workflow
Once your project has been setup, import the WDL workflow 02_download_collection_files.wdl to the DNAnexus platform using dxCompiler.
| |
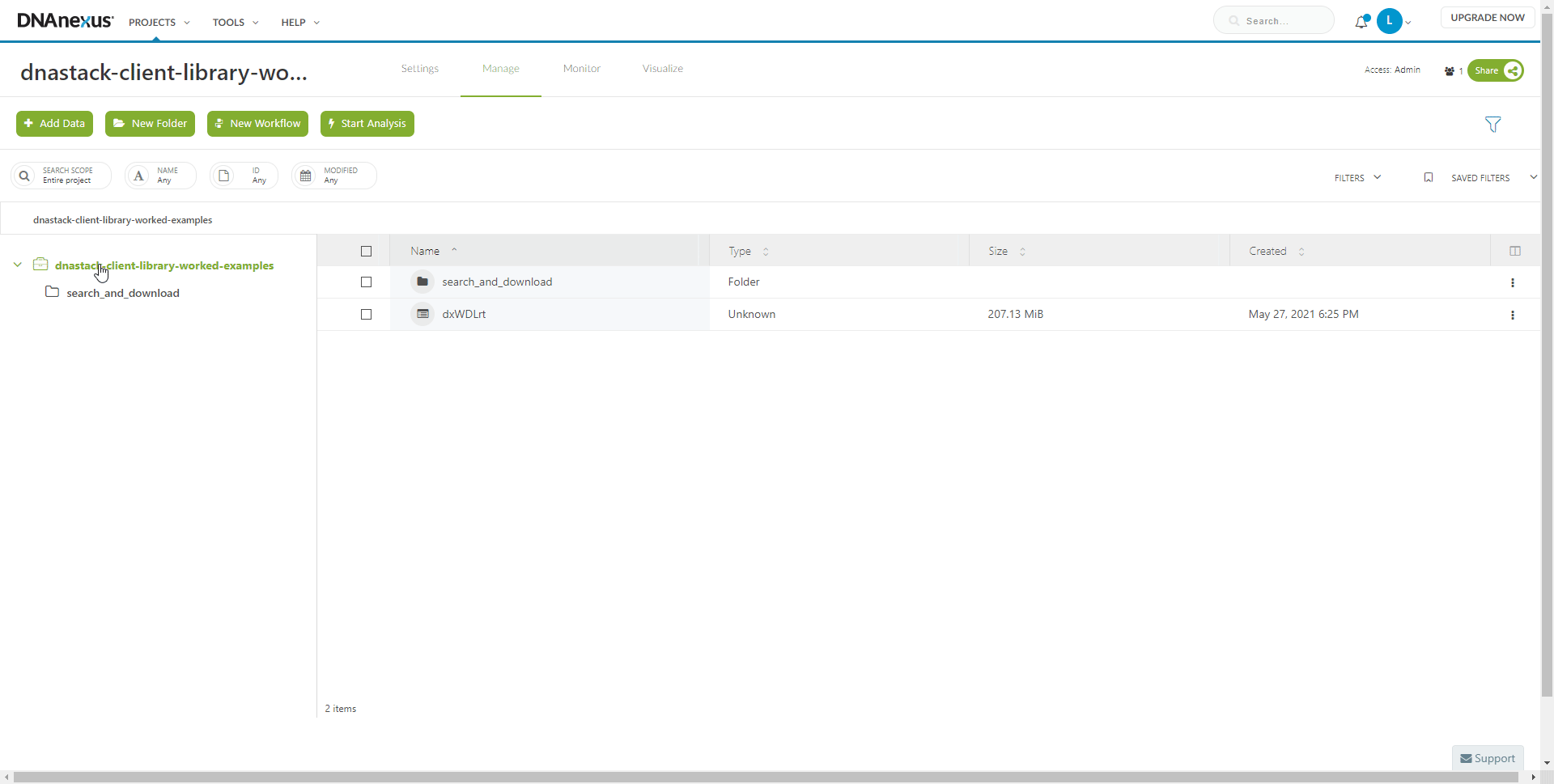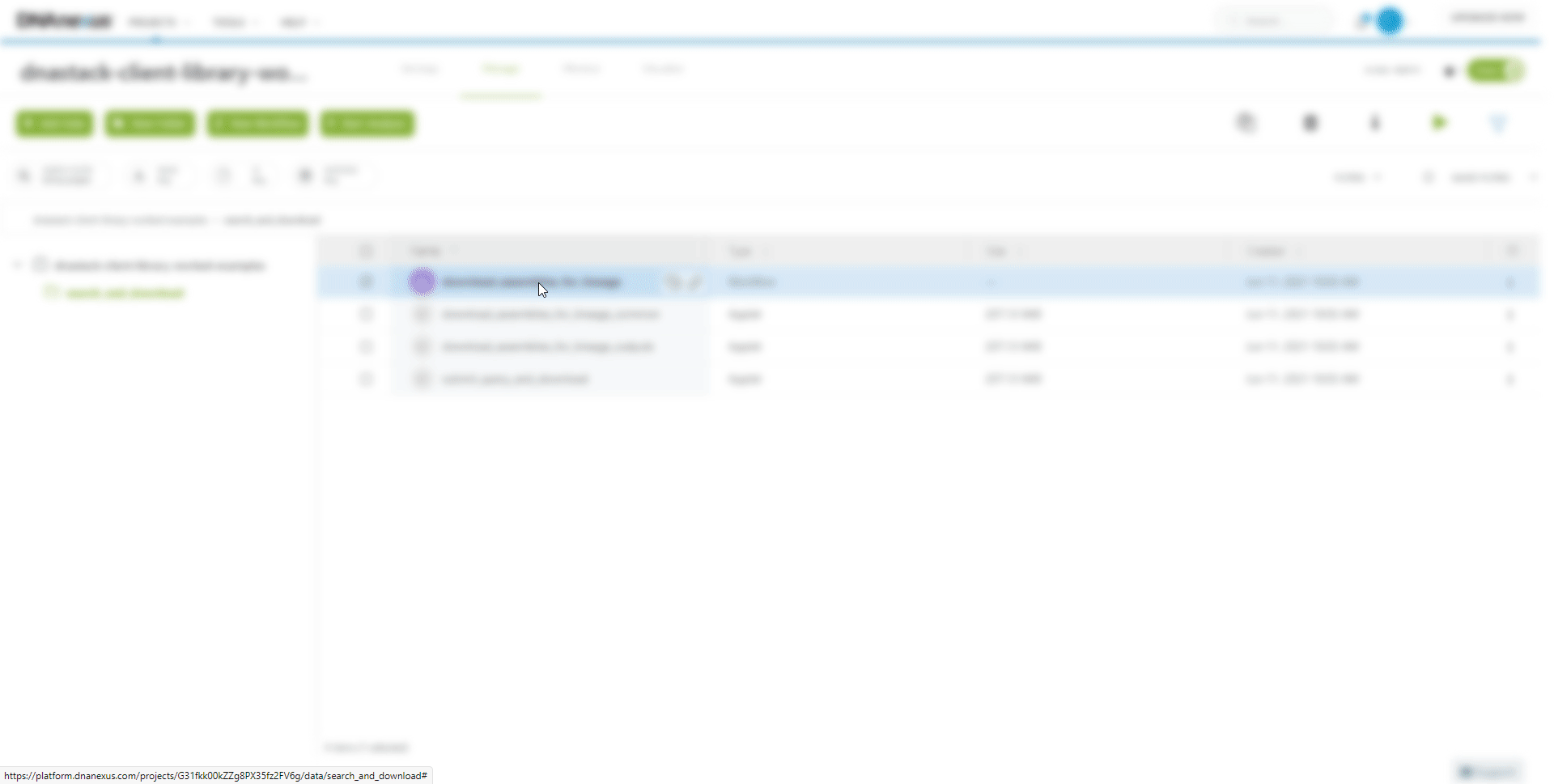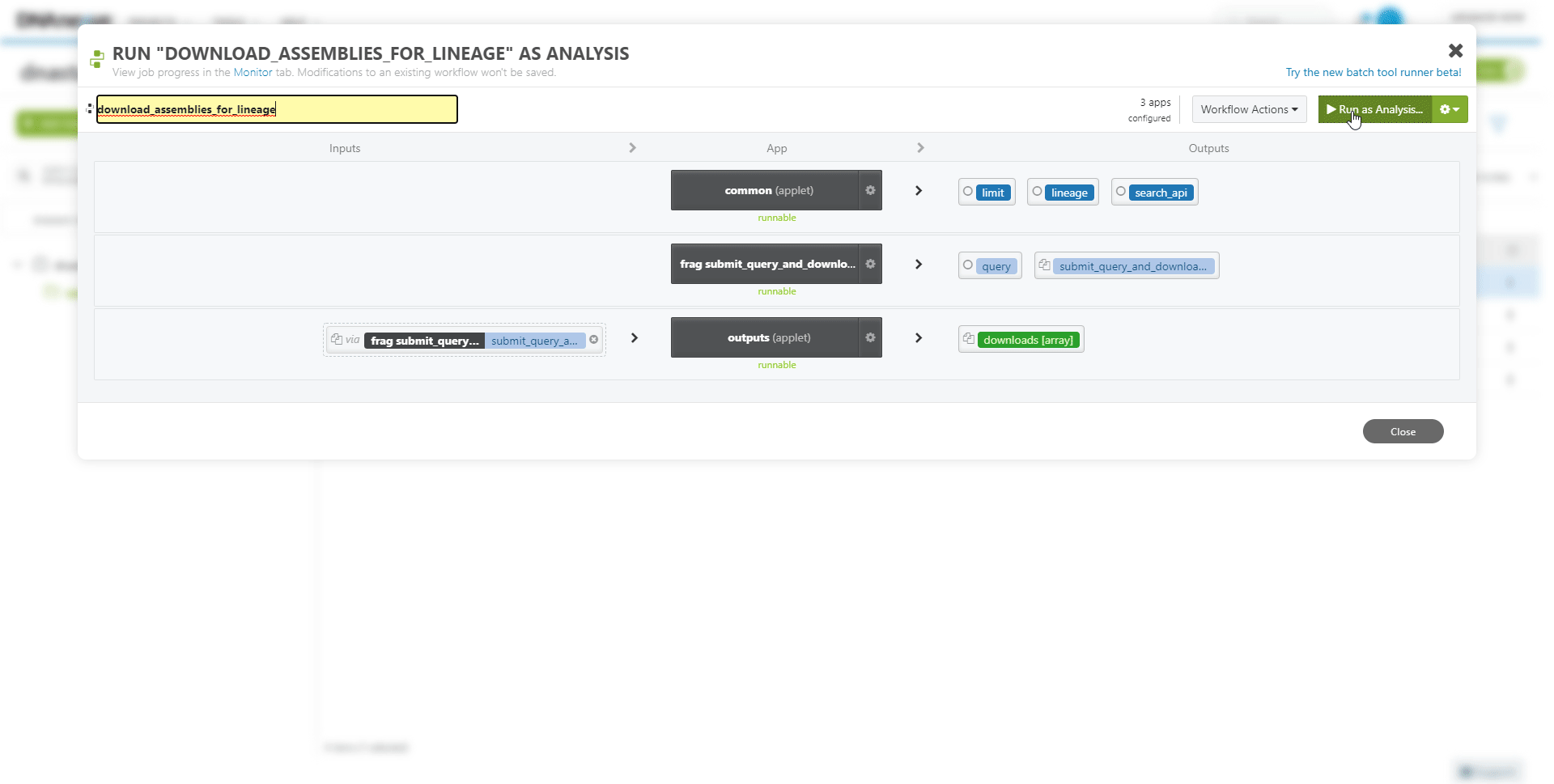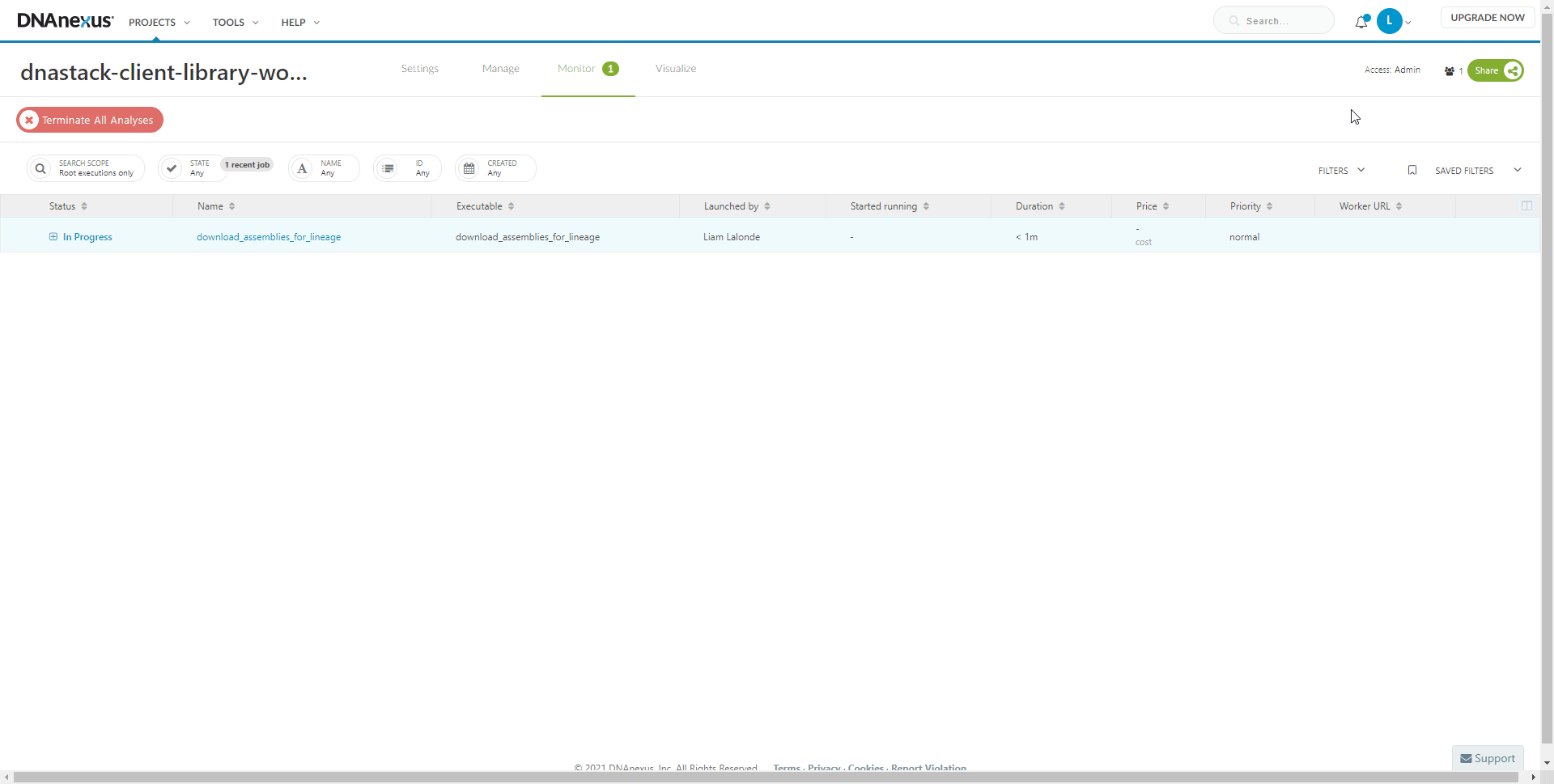
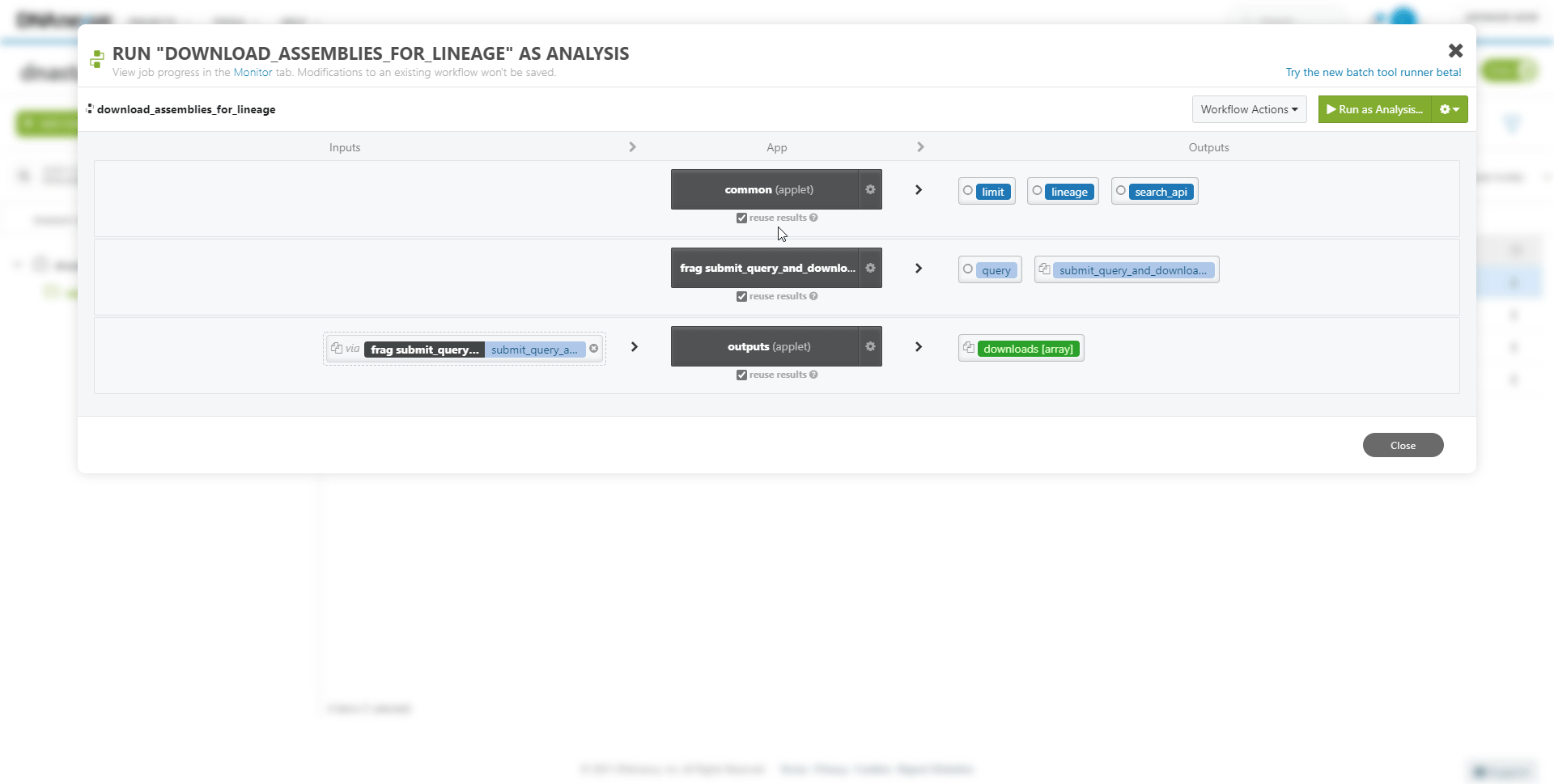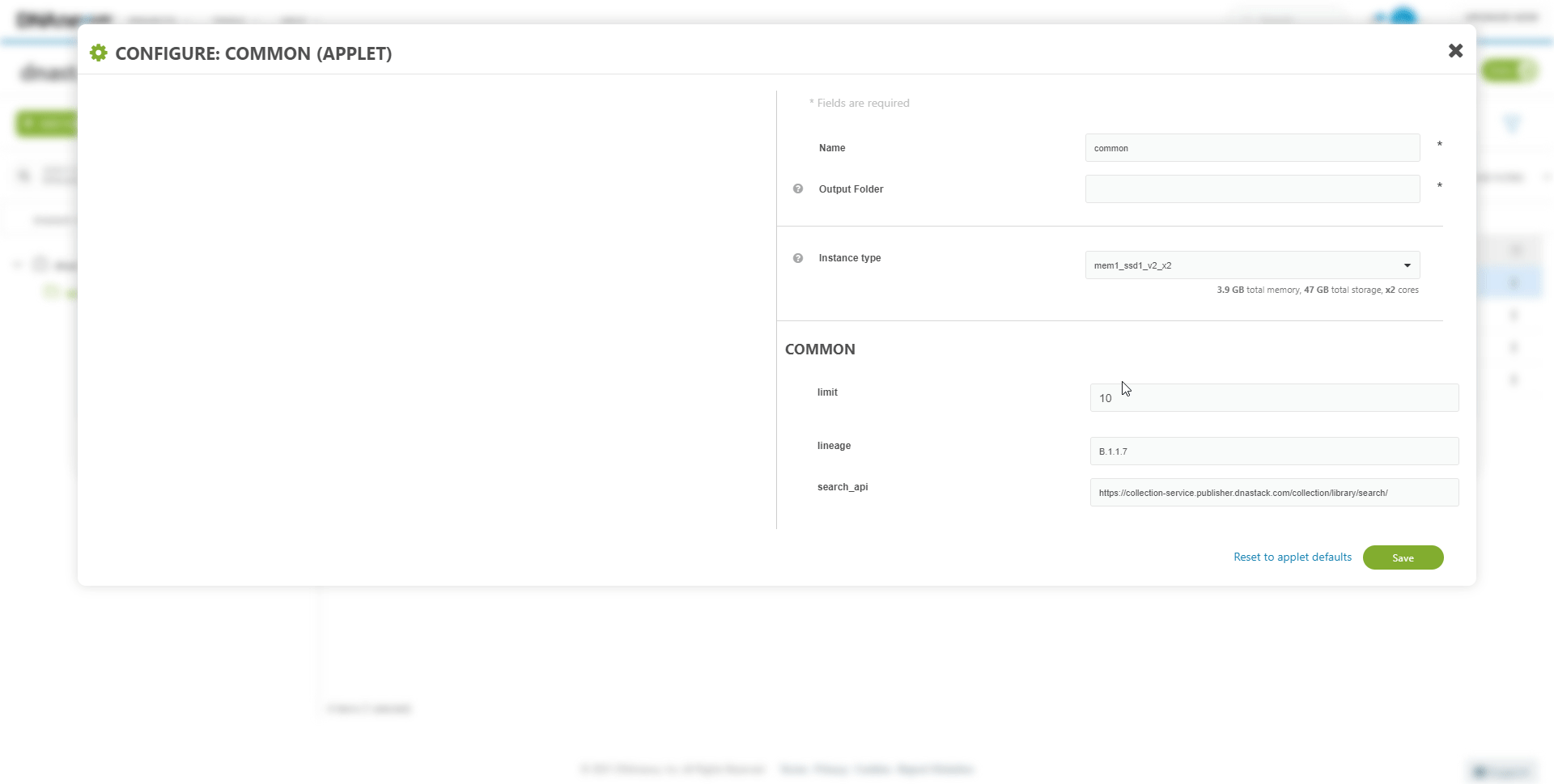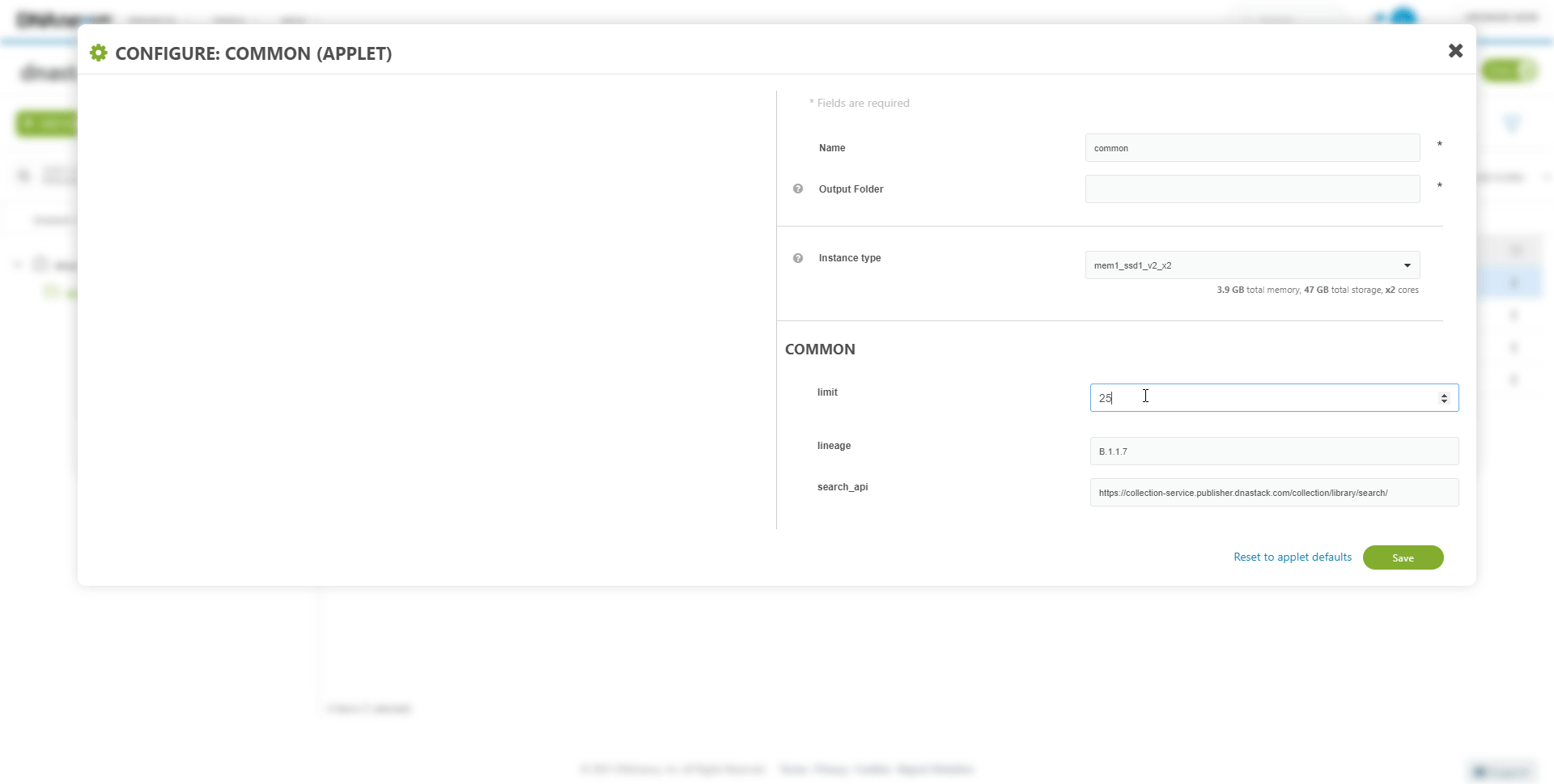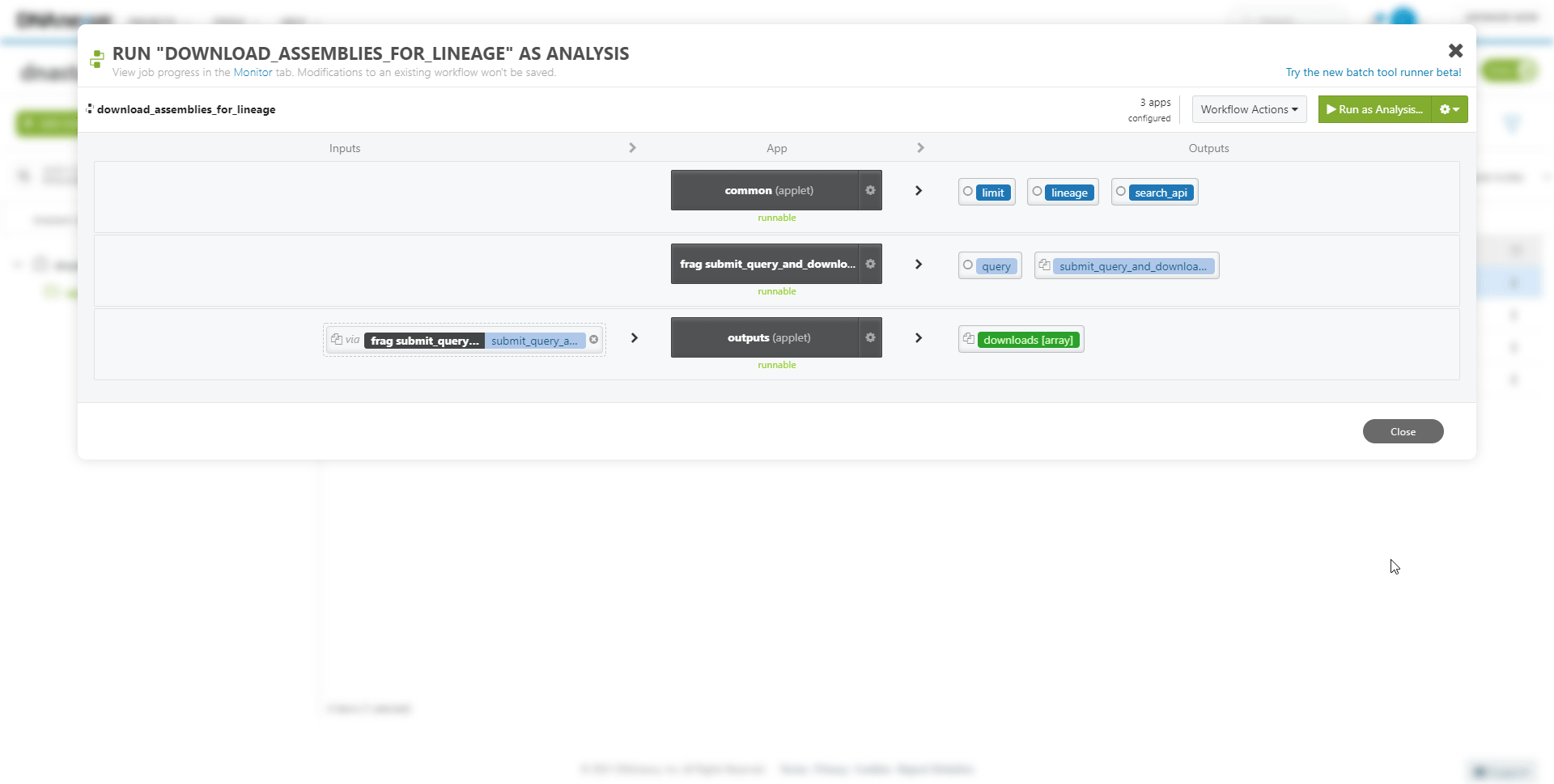
To run the newly created workflow on the DNAnexus platform: Select the project folder and Workflow item, and press Run as Analysis.
To view the progress of the workflow at any time: select the Monitor tab.
Inputs or parameters of the workflow can be modified. For example, to download from a different collection.
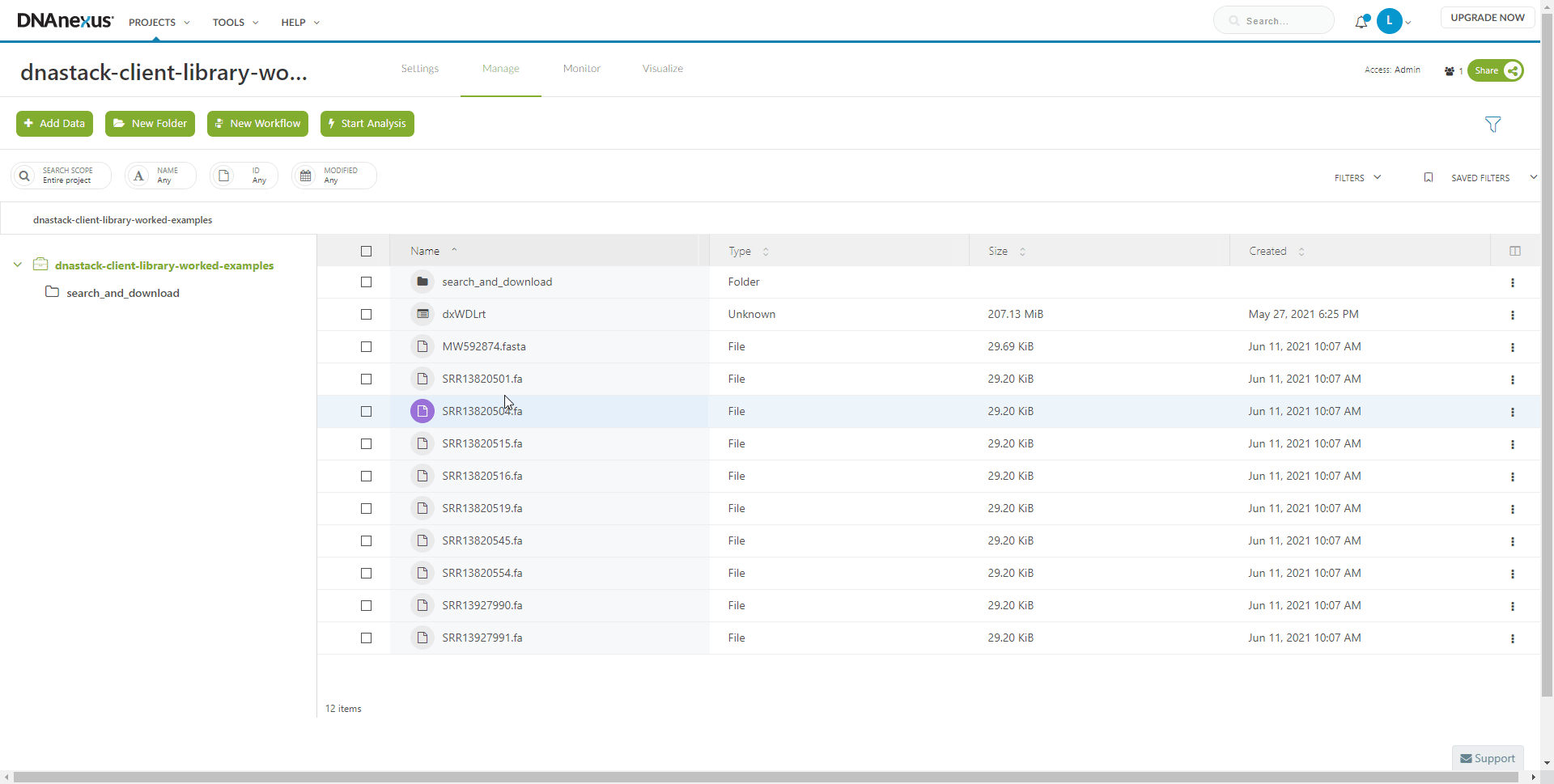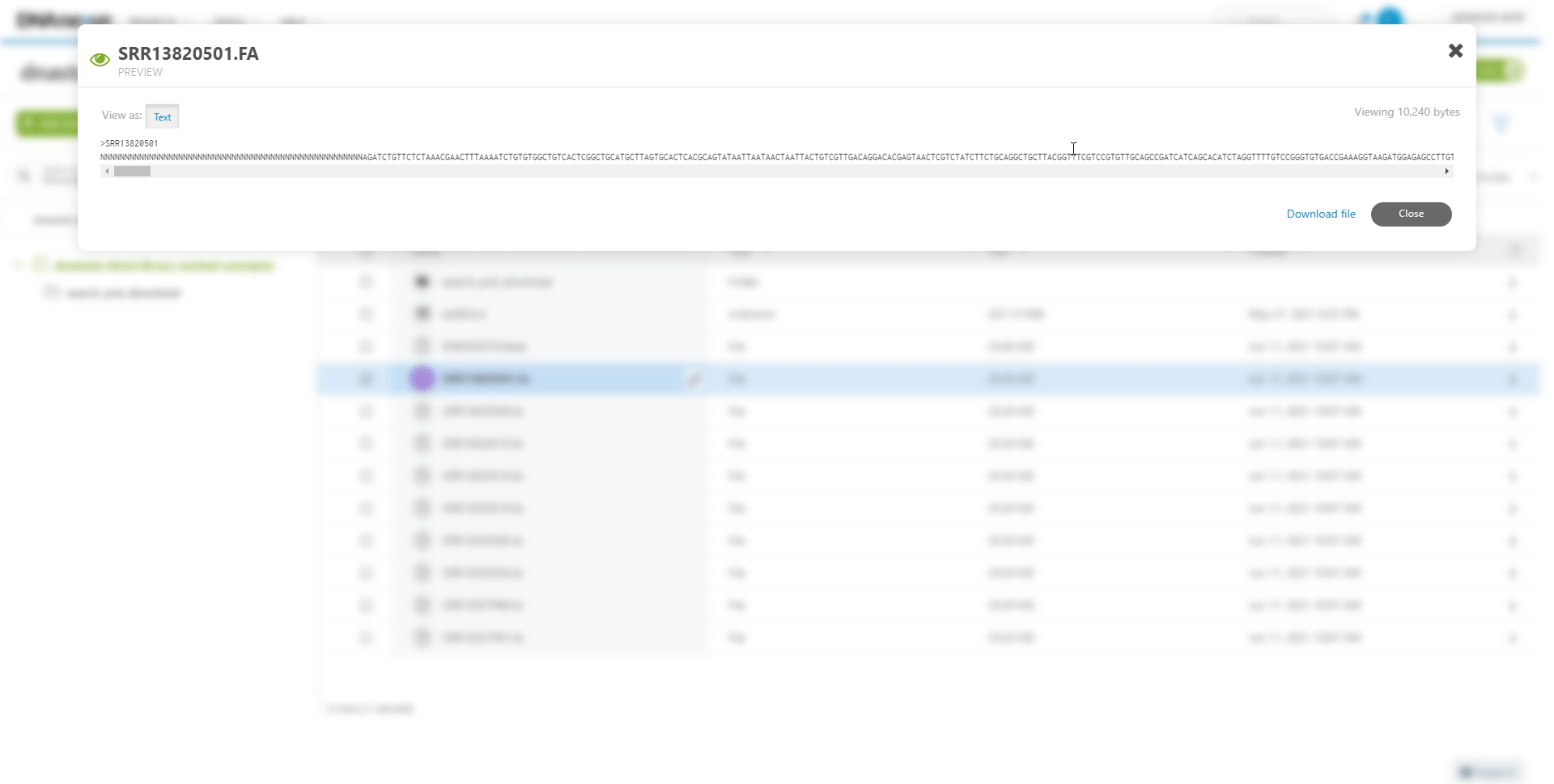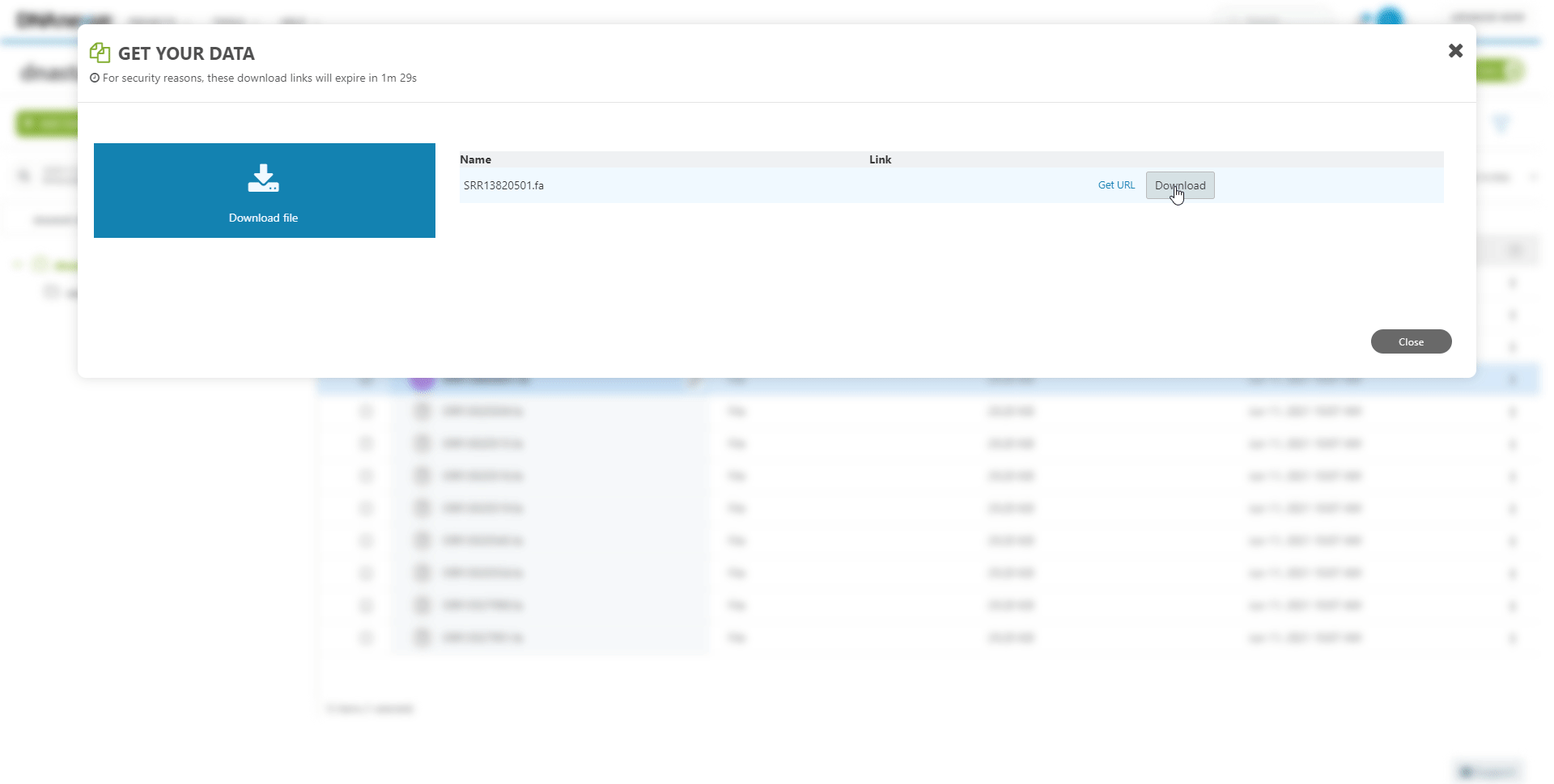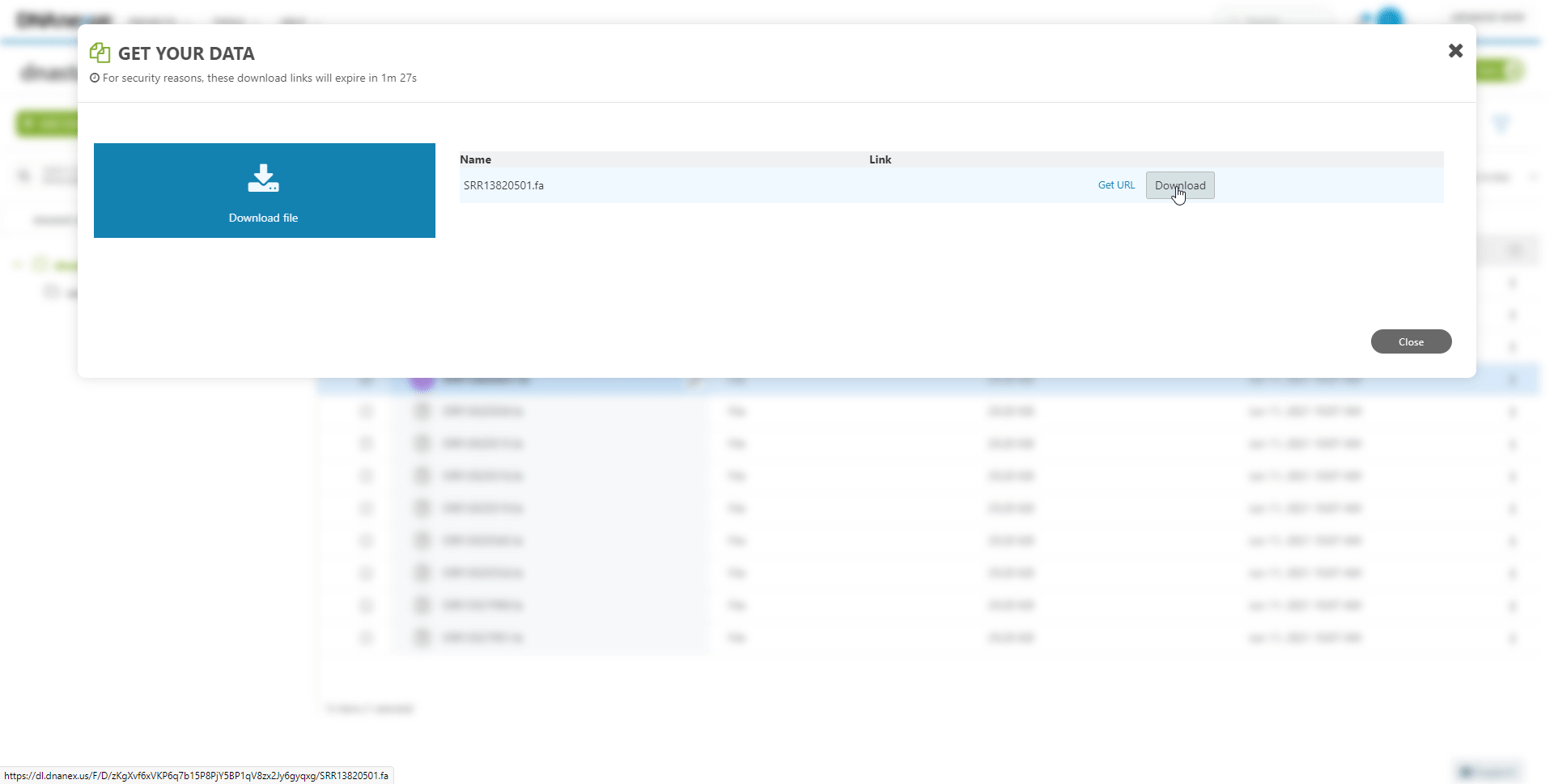
The output files can be found in and downloaded from the folder specified in the Run dialog. In this case, the root of the DNAnexus project.